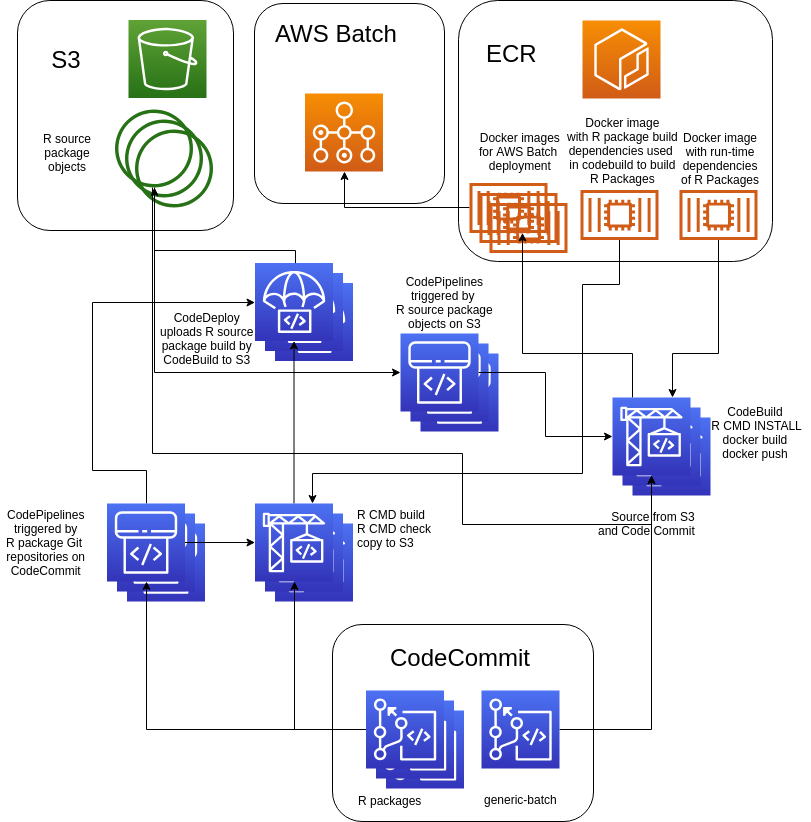
CI/CD enabled architecture for cloud data science platform
Background For a work project, the backend of a life science related data science platform had to be re-engineerd towards higher modularity. The aim was to simplify the integration of new algorithms in the platform and reducing the amount of repated code.
Some requirements were given by the interfacing components. There existed a web app that allows uploading data into a cloud object store and processing it using a container based process queue in the cloud. Container recipe and data processing scripts in R were kept together in git repositories.
For the redesign, best practices for R were implemented, the most significant change was to re-write scripts into well-defined R packages. Further, separate cloud CI/CD pipelines for R packages and packaging of Docker images were setup.
As shown in the below chart, AWS cloud services were used.